Primer Definition, Primer Design, Types, Online Tools, Uses
Table Of Content
The number of consecutive Gs and Cs at the 3' end of both the left and right primer.
New to the PrimerQuest Tool?
Here, we used SADDLE to design a set of 60 primers to detect 56 actionable gene fusions for non-small cell lung cancer (NSCLC) across six genes (ALK, ROS1, RET, NRTK1, NTRK2, and NRTK3). The number of primers are lower than 56 × 2 because the same exon can be fused with multiple partner genes or exons. We detect the fusions in complementary DNA (cDNA) reverse transcribed from RNA, in order to limit the complexity and length of the detection targets. For each fusion of interest, the primer set includes a forward primer (fP) targets the upstream partner gene and a reverse primer (rP) targets the downstream partner gene (Fig. 5a).
Primer Design using Gibson Cloning Method
However, primers do not need to correspond to the template strand completely; it is essential, however, that the 3’ end of the primer corresponds completely to the template DNA strand so elongation can proceed. Usually a guanine or cytosine is used at the 3’ end, and the 5’ end of the primer usually has stretches of several nucleotides. Also, both of the 3’ ends of the hybridized primers must point toward one another. N.G.X., M.X.W., and D.Y.Z. wrote the program for primer design and data analysis.
High-throughput primer design by scoring in piecewise logistic model for multiple polymerase chain reaction variants
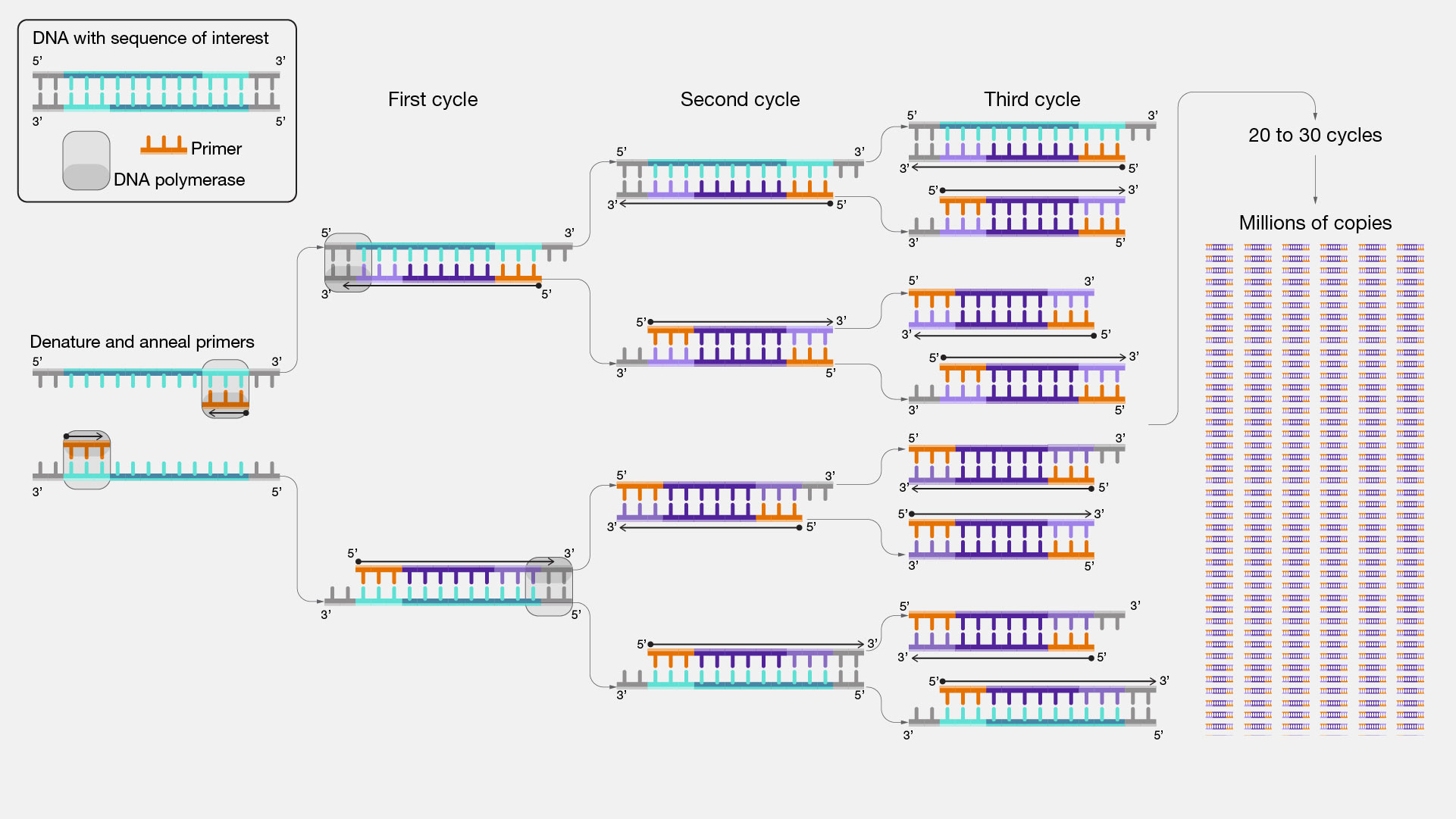
In PCR, both forward and reverse primers are used together to amplify the desired DNA region. The forward primer binds to the antisense strand, and the reverse primer binds to the positive-sense strand, flanking the target DNA sequence. This allows DNA polymerase to synthesize new DNA strands in both directions from the primers, resulting in the amplification of the specific DNA region of interest. In summary, DNA primers are short DNA sequences used in PCR to target specific DNA regions for amplification and subsequent analysis. They serve as the starting point for DNA synthesis and are essential for the success of the PCR process.
Haplotyping pharmacogenes using TLA combined with Illumina or Nanopore sequencing Scientific Reports - Nature.com
Haplotyping pharmacogenes using TLA combined with Illumina or Nanopore sequencing Scientific Reports.
Posted: Sat, 22 Oct 2022 07:00:00 GMT [source]
Alternatively, it can simplify the workflow of moderate size NGS and qPCR assays by removing the need for strict contamination control from open-tube steps. Data can be uploaded into PrimerMapper from a local file or from NCBI’s GenBank22 or dbSNP23 databases. SNP sequence data is preprocessed by PrimerMapper prior to execution to collect the SNP position and type from the header field and also to remove whitespace from the header field so as to preserve the entire sequence name. The user sets the various parameters for primer design for DNA sequence or SNP sequences such as primer maximum and minimum lengths, GC%, melting temperature (Tm), etc.

Primers can be visualized as arrows or lines and also a ‘magnifying glass’ can be checked so as to magnify the primer names by moving the mouse over the primers within the primer-map canvas (circle 2). The user can also get the base pair position along the sequence (circle 3) by moving the mouse under the sequence map. The user can also draw new primers on the canvas (circle 4) which will return a JavaScript popup box with the DNA sequence, GC% and primer Tm values of the sequence corresponding to the line drawn by the user (circles 4 and 5).
Regional websites
Multiplex PCR products and ligation products were all purified using DNA Clean & Concentrator Kits (ZYMO Research). The volume of DNA-binding buffer was 250 μl for multiplex PCR products clean-up, and 482.5 μl for ligation products clean-up; 25 μl Milli-Q water was used as elution buffer for each reaction. End repair mixture was ligated with adapters using NEBNext® Ultra™ II Ligation Module (New England Biolabs). Each reaction was a mixture of 30 μl NEBNext Ultra II Ligation Master Mix, 1 μl NEBNext Ligation Enhancer, 2.5 μl NEBNext Adaptor for Illumina, and 60 μl previous End repair mixture. Thermocycling started with the incubation at 20 °C for 15 min with the heated lid off; after adding 3 μl USER™ enzyme to the ligation mixture, the reaction was incubated at 37 °C for 15 min with the heated lid set to 55 °C.
Can my primer amplify if the forward is located in an intron region? - ResearchGate
Can my primer amplify if the forward is located in an intron region?.
Posted: Sat, 18 Jan 2014 08:00:00 GMT [source]
Primer- Definition, Types, Primer Design Online Tools, Uses
We begin our implementation of primer candidate generation through the selection of one or more “pivot” nucleotides on human genomic DNA around which we design the forward and reverse primers (Fig. 2a). The pivot nucleotides are the ones that must be included in the amplicon insert, and for example could be the hotspot region of a gene that is frequently mutated. From the pivot nucleotides and a constraint on the maximum length of the amplicons (e.g., determined by the read length of NGS), we can systematically generate a series of different proto-primers with 3′ end just outside the pivot nucleotides. The proto-primers have a large range of different lengths and binding energies to their complementary sequences, and will next be trimmed at the 3′ end to generate the primer candidates (Fig. 2a). Enabling this option will make it much easier to find gene-specific primers since there is no need to distinguish between splice variants. This option requires you to enter a refseq mRNA accession or gi or fasta sequence as PCR template input because other type of input may not allow the program to properly interpret the result.
Can RNA primers be used in PCR?
Moreover, the DNA binding sequence of the primer in vitro has to be specifically chosen, which is done using a method called basic local alignment search tool (BLAST) that scans the DNA and finds specific and unique regions for the primer to bind. With this option on, the program will search the primers against the selected database and determine whether a primer pair can generate a PCR product on any targets in the database based on their matches to the targets and their orientations. The program will return, if possible, only primer pairs that do not generate a valid PCR product on unintended sequences and are therefore specific to the intended template. Note that the specificity is checked not only for the forward-reverse primer pair, but also for forward-forward as well as reverse-reverse primer pairs.
For this reason, degenerate primers are also used when primer design is based on protein sequence, as the specific sequence of codons are not known. Therefore, primer sequence corresponding to the amino acid isoleucine might be "ATH", where A stands for adenine, T for thymine, and H for adenine, thymine, or cytosine, according to the genetic code for each codon, using the IUPAC symbols for degenerate bases. Degenerate primers may not perfectly hybridize with a target sequence, which can greatly reduce the specificity of the PCR amplification. We next examined the top five most dominant Dimer reads in the library (Fig. 4d) and compared them to the top five predicted dimer reads based on the Badness function (Fig. 4e).
C(g) is a function that is monotonically non-increasing in g, indicating decreasing tolerance to detrimental changes at later generations. The parameter gt indicates the generation in which simulated annealing terminates, and we switch over to stochastic gradient descent. Useful tips to keep in mind when designing your seamless cloning projects.

However, both steps are labor-intensive and cannot be applied universally to all multiplexed PCR reactions. In contrast, relatively little systematic work has been reported on computational approaches to minimizing the formation of primer dimers in the first place. To the best of our knowledge, existing multiplex primer design algorithm never exceeded 70 primer pairs in one tube11,12,13,14. This is mainly due to the high computational cost when the number of primers increases15.
Importantly, library preparation using optimized primer sets generated by SADDLE does not depend on labor-intensive enzymatic cleavage or size selection steps to remove dimers. The diversification of PCR-based methodologies coupled with a rapid expansion in available genomic data has put further demands on the scale and utility of primer design software. In particular, batch primer design has become an important feature of primer design tools to accommodate screening large numbers of genes and datasets.
Comments
Post a Comment